About Us
Dr. Gregory is the Margaret Harris and David Silverman Distinguished Professor of Neurosurgery, the Vice Chair of Research in the Department of Neurology, and Director of the Molecular Genomics Core at the Duke Molecular Physiology Institute. As a neurogenomicist, Dr. Gregory applies the experience gained from leading the sequencing of chromosome 1 for the Human Genome Project to elucidating the mechanisms underlying multi-factorial diseases using genetic, genomic, and epigenetic approaches. Dr. Gregory’s primary areas of research involve understanding the molecular processes associated with disease development and progression of multiple sclerosis (MS) and other complex diseases. His research includes the investigation of a novel drug in inducing white matter injury repair and the characterization of lesion microenvironmental changes in MS. Dr. Gregory is the Principal Investigator of the MURDOCK-MS collection, a cross sectional MS cohort of ~1000 MS patients that provide the basis for genetic, genomic and metabolomic biomarker identification of MS disease development and progression. Dr. Gregory is the Research Director of the Duke Center of Research in Autoimmunity and MS (DREAMS) within the Department of Neurology.
Research
The Gregory lab is involved in several lab-based and collaborative research projects that focus on identifying the genomic, genetic, and epigenetic underpinnings of complex disease.
Multiple Sclerosis:
In 2007 Dr Gregory and his collaborators identified the first MS gene outside of the MHC to be associated with the disease (1). The finding forms the basis of ongoing functional research to identify the mechanism by which the cytokine receptor (IL7R) and ILR are implicated in the disease. Dr. Gregory's lab is assessing T cell signaling of IL7 in MS patients, understanding the mechanism of IL7R splicing (2), and construction of an IL7R mouse model of the disease in collaboration with Dr. Mariano Garcia Blanco (UTMB). Dr. Gregory is Principal Investigator of the MS-MURDOCK study. The study has developed a ~1,000 patient multiple sclerosis collection that is independent of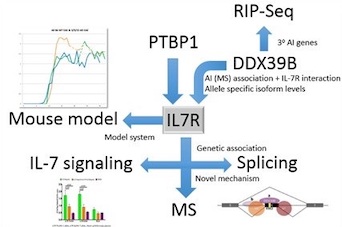 subtype with the aims of understanding the mechanisms associated with MS development and progression, and the generation of multi-omic biomarkers to facilitate reclassification of the disease. Finally, Dr. Gregory is using a mouse model of MS experimental autoimmune encephalomyelitis (EAE), to understand the molecular mechanisms of disease development and efficacy of novel drugs. The Gregory lab is exploring the dynamics of mRNA and miRNA expression during EAE course using next generation sequencing approaches to elucidate the changes and interrelationships of gene function during EAE development and recovery. These data will provide the baseline for evaluating the mechanism of novel cyclotide drug function in collaboration with Dr. Christian Gruber (University of Brisbane). (3), and hydroxyl-oxysterols with Drs. Eric Benner (Duke, Pediatrics) and Mari Shinohara (Duke, Immunology).
subtype with the aims of understanding the mechanisms associated with MS development and progression, and the generation of multi-omic biomarkers to facilitate reclassification of the disease. Finally, Dr. Gregory is using a mouse model of MS experimental autoimmune encephalomyelitis (EAE), to understand the molecular mechanisms of disease development and efficacy of novel drugs. The Gregory lab is exploring the dynamics of mRNA and miRNA expression during EAE course using next generation sequencing approaches to elucidate the changes and interrelationships of gene function during EAE development and recovery. These data will provide the baseline for evaluating the mechanism of novel cyclotide drug function in collaboration with Dr. Christian Gruber (University of Brisbane). (3), and hydroxyl-oxysterols with Drs. Eric Benner (Duke, Pediatrics) and Mari Shinohara (Duke, Immunology).
Autism:
Recent CDC estimates suggest that autism affects more than one in 68 children in the US. The Gregory lab is using independent approaches to not only understand the genetic and epigenetic mechanisms underlying autism, but also how children can be treated to resolve their symptoms. Dr. Gregory is project PI in the SOARS-B consortium headed by Dr. Lin Sikich of Duke University's Center for Autism and Brain Development. This exciting new clinical trial is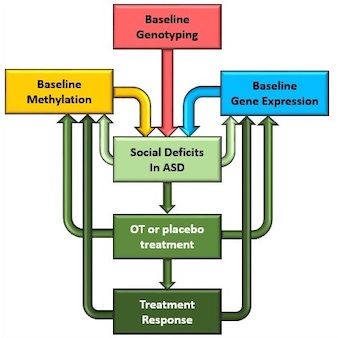 assessing the efficacy of nasally delivered oxytocin to ameliorate some of the core deficits of autism. The Gregory lab's role in the consortium is to develop genetic and epigenetic predictors of oxytocin response and to assess the long term effects of drug exposure on these modalities (4). In collaboration with Drs. Sheryl Moy (UNC, Psychiatry) and Dr. Yong-hui Jiang (Duke, Pediatrics), Dr. Gregory's lab has recently been awarded an NIH grant to explore the mechanisms of oxytocin response in an animal model of the disease, to extend the epigenetic profiling of SOAR-B responders, and to refine the epigenetic regulation of the oxytocin receptor (OXTR). The findings of this grant will provide valuable data for the mode of action of oxytocin response in specific regions of the brain that will applicable to clinical trials of oxytocin response in numerous psychosocial phenotypes, including autism. Finally, together with Professor Emeritus of Pediatrics Dr. G. Robert Delong, Dr. Gregory is investigating how epigenetic factors within a multigenerational family can lead to the development of the disorder and how the identification of compound genetic risk factors in psychosocial families by exome sequencing may lead to the development of autism.
assessing the efficacy of nasally delivered oxytocin to ameliorate some of the core deficits of autism. The Gregory lab's role in the consortium is to develop genetic and epigenetic predictors of oxytocin response and to assess the long term effects of drug exposure on these modalities (4). In collaboration with Drs. Sheryl Moy (UNC, Psychiatry) and Dr. Yong-hui Jiang (Duke, Pediatrics), Dr. Gregory's lab has recently been awarded an NIH grant to explore the mechanisms of oxytocin response in an animal model of the disease, to extend the epigenetic profiling of SOAR-B responders, and to refine the epigenetic regulation of the oxytocin receptor (OXTR). The findings of this grant will provide valuable data for the mode of action of oxytocin response in specific regions of the brain that will applicable to clinical trials of oxytocin response in numerous psychosocial phenotypes, including autism. Finally, together with Professor Emeritus of Pediatrics Dr. G. Robert Delong, Dr. Gregory is investigating how epigenetic factors within a multigenerational family can lead to the development of the disorder and how the identification of compound genetic risk factors in psychosocial families by exome sequencing may lead to the development of autism.
Cardiovascular Disease:
It is estimated that every one in four deaths in the US is attributable to heart disease and the health burden is believed to be greater than $100 billion annually. Dr. Gregory is collaborating with Drs. Svati Shah, Bill Kraus, and Elizabeth Hauser to identify the genetic architecture of the disease using Duke's unique CATHGEN cohort via GWAS and candidate gene association studies, metabolomic profiling with Dr. Chris Newgard, and transcriptomic and epigenomic approaches (5,6,7). The latter, profiling the methylome of cardiovascular disease, also forms the basis of collaboration with cardiologists Drs. Svati Shah, Asad Shah, and G. Chad Hughes to identify DNA methylation and gene expression differences during bi- and tricuspid aorta development.
Elizabeth Hauser to identify the genetic architecture of the disease using Duke's unique CATHGEN cohort via GWAS and candidate gene association studies, metabolomic profiling with Dr. Chris Newgard, and transcriptomic and epigenomic approaches (5,6,7). The latter, profiling the methylome of cardiovascular disease, also forms the basis of collaboration with cardiologists Drs. Svati Shah, Asad Shah, and G. Chad Hughes to identify DNA methylation and gene expression differences during bi- and tricuspid aorta development.
Developmental Projects:
The Gregory lab is involved in a number of high-risk high-reward projects to identify the modalities of disease development. In collaboration with Dr. Chris Newgard, the Gregory lab is profiling gene expression changes in pancreatic islets under the regulation of Pdx and Nkx in a rat model of diabetes using next generation RNA-, ChIP-Seq and single cell expression profile approaches; while a collaboration with Drs. Fashid Guilak (Wash. U) and Dianne Little (Duke, Orthopedics) is aimed at identifying the epigenetic mechanisms associated with diet induced changes of mouse stem cells in the development of obesity and OA
Recent Publications
Skewing of the population balance of lymphoid and myeloid cells by secreted and intracellular osteopontin. Kanayama M, Xu S, Danzaki K, Gibson JR, Inoue M, Gregory SG, Shinohara ML. Nat Immunol. 2017 Jul 3. doi: 10.1038/ni.3791.
Whole blood sequencing reveals circulating microRNA associations with high-risk traits in non-ST-segment elevation acute coronary syndrome. Wang A, Kwee LC, Grass E, Neely ML, Gregory SG, Fox KAA, Armstrong PW, White HD, Ohman EM, Roe MT, Shah SH, Chan MY. Atherosclerosis. 2017 Jun;261:19-25. doi: 10.1016/j.atherosclerosis.2017.03.041. Epub 2017 Mar 30.
Genome-wide association study identifies three novel loci in Fuchs endothelial corneal dystrophy. Afshari NA, Igo RP Jr, Morris NJ, Stambolian D, Sharma S, Pulagam VL, Dunn S, Stamler JF, Truitt BJ, Rimmler J, Kuot A, Croasdale CR, Qin X, Burdon KP, Riazuddin SA, Mills R, Klebe S, Minear MA, Zhao J, Balajonda E, Rosenwasser GO, Baratz KH, Mootha VV, Patel SV, Gregory SG, Bailey-Wilson JE, Price MO, Price FW Jr, Craig JE, Fingert JH, Gottsch JD, Aldave AJ, Klintworth GK, Lass JH, Li YJ, Iyengar SK. Nat Commun. 2017 Mar 30;8:14898. doi: 10.1038/ncomms14898.
A genome-wide trans-ethnic interaction study links the PIGR-FCAMR locus to coronary atherosclerosis via interactions between genetic variants and residential exposure to traffic. Ward-Caviness CK, Neas LM, Blach C, Haynes CS, LaRocque-Abramson K, Grass E, Dowdy ZE, Devlin RB, Diaz-Sanchez D, Cascio WE, Miranda ML, Gregory SG, Shah SH, Kraus WE, Hauser ER. PLoS One. 2017 Mar 29;12(3):e0173880. doi: 10.1371/journal.pone.0173880. eCollection 2017.
Human Epistatic Interaction Controls IL7R Splicing and Increases Multiple Sclerosis Risk. Galarza-Muñoz G, Briggs FB, Evsyukova I, Schott-Lerner G, Kennedy EM, Nyanhete T, Wang L, Bergamaschi L, Widen SG, Tomaras GD, Ko DC, Bradrick SS, Barcellos LF, Gregory SG, Garcia-Blanco MA. Cell. 2017 Mar 23;169(1):72-84.e13. doi: 10.1016/j.cell.2017.03.007
An interferon-β-resistant and NLRP3 inflammasome-independent subtype of EAE with neuronal damage. Inoue M, Chen PH, Siecinski S, Li QJ, Liu C, Steinman L, Gregory SG, Benner E, Shinohara ML.Nat Neurosci. 2016 Dec;19(12):1599-1609. doi: 10.1038/nn.4421. Epub 2016 Nov 7.
Human centromere repositioning within euchromatin after partial chromosome deletion.Sullivan LL, Maloney KA, Towers AJ, Gregory SG, Sullivan BA.Chromosome Res. 2016 Dec;24(4):451-466. Epub 2016 Aug 31.
Case-Only Survival Analysis Reveals Unique Effects of Genotype, Sex, and Coronary Disease Severity on Survivorship.Dungan JR, Qin X, Horne BD, Carlquist JF, Singh A, Hurdle M, Grass E, Haynes C, Gregory SG, Shah SH, Hauser ER, Kraus WE.PLoS One. 2016 May 17;11(5):e0154856. doi: 10.1371/journal.pone.0154856. eCollection 2016.
Genetic Variants in the Bone Morphogenic Protein Gene Family Modify the Association between Residential Exposure to Traffic and Peripheral Arterial Disease.Ward-Caviness CK, Neas LM, Blach C, Haynes CS, LaRocque-Abramson K, Grass E, Dowdy E, Devlin RB, Diaz-Sanchez D, Cascio WE, Lynn Miranda M, Gregory SG, Shah SH, Kraus WE, Hauser ER.PLoS One. 2016 Apr 15;11(4):e0152670. doi: 10.1371/journal.pone.0152670. eCollection 2016.
Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis.Kraus WE, Muoio DM, Stevens R, Craig D, Bain JR, Grass E, Haynes C, Kwee L, Qin X, Slentz DH, Krupp D, Muehlbauer M, Hauser ER, Gregory SG, Newgard CB, Shah SH.PLoS Genet. 2015 Nov 5;11(11):e1005553. doi: 10.1371/journal.pgen.1005553. eCollection 2015 Nov.
Our Team
Former Lab Members
Rachel Cote PhD (UNC Chapel Hill)
Christina Sheedy PhD (Duke UPGG Program)
Matthew Schemmel (Illumia)
Josh Virgadamo (U.S. Navy)
Jennifer Doss (Duke UPGG Program)
Shera Watson (Duke Cardiology)
Aaron Towers (Duke UPGG Program)
Former Trainees
Mollie Minear PhD - UPGG Graduate Program (Fellow - NIH, National heart, Lung, and Blood Institute)
Christina Markunas PhD - UPGG Graduate Program (RTI - Epigenetic Epidemiologist)
Jessica Connelly PhD - Post Doctoral Fellow (University of Virginia - Faculty)
Beth Sutton PhD - Post Doctoral Fellow (Campbell University - Faculty)
Jama Purser MD - Clinical Fellow
Current and Past Undergraduate Trainees
Sofia Velasquez – Colgate University
Nicole Joy - Duke University
Lauren Vaughan - Duke University
Angeline Luong - Duke University
Hunter Nisonoff - Duke University
Thomas Boyle - Duke University
Allison Dorogi - Duke University
Jaret (Mac) Karnuta - Duke University
Charles Zhao – Duke University
Nava Barman - Duke University
Cynthia Rouf – North Carolina State University
William Morgenlander – Notre Dame
Aisha Venugopal - UNC, Chapel Hill
Seth Newman - Washington University
Current and Past High School Trainees
Maya Watson – Durham Academy
Grace Mott – Durham School of the Arts
Kaitlyne Sheehan – Durham School of the Arts
Maya Montani - Durham School of the Arts
Reuben Tacas – Durham School of the Arts
Sam Finlay – Durham School of the Arts
Drew Harrelson – North Carolina School of Science and Math
Hailey Gosnell – North Carolina School of Science and Math
Anna Scotton – North Carolina School of Science and Math
Jamie Chamberlin – North Carolina School of Science and Math








